I was trying to simulate "Sampling Distribution of Sample Proportions" using Python. I tried with a Bernoulli Variable as in example here
The crux is that, out of large number of gumballs, we have yellow balls with true proportion of 0.6. If we take samples (of some size, say 10), take mean of that and plot, we should get a normal distribution.
I tried to do in python but I only always get uniform distribution (or flats out in the middle). I am not able to understand what am I missing.
Program:
from SDSP import create_bernoulli_population, get_frequency_df
from random import shuffle, choices
from bi_to_nor_demo import get_metrics, bare_minimal_plot
import matplotlib.pyplot as plt
N = 10000 # 10000 balls
p = 0.6 # probability of yellow ball is 0.6, and others (1-0.6)=>0.4
n_pickups = 1000 # sample size
n_experiments = 100 # I dont know what this is called
# generate population
population = create_bernoulli_population(N,p)
theor_df = get_frequency_df(population)
theor_df
# choose sample, take mean and add to X_mean_list. Do this for n_experiments times
X_hat = []
X_mean_list = []
for each_experiment in range(n_experiments):
X_hat = choices(population, k=n_pickups) # this method is with replacement
shuffle(population)
X_mean = sum(X_hat)/len(X_hat)
X_mean_list.append(X_mean)
# plot X_mean_list as bar graph
stats_df = get_frequency_df(X_mean_list)
fig, ax = plt.subplots(1,1, figsize=(5,5))
X = stats_df['x'].tolist()
P = stats_df['p(x)'].tolist()
ax.bar(X, P, color="C0")
plt.show()
Dependent functions:
bi_to_nor_demo
SDSP
Update: I even tried uniform distribution as below but getting similar output. Not converging to normal :(. (using below function in place of create_bernoulli_population)
def create_uniform_population(N, Y=[]):
"""
Given the total size of population N,
this function generates list of those outcomes uniformly distributed
population list
N - Population size, eg N=10000
p - probability of interested outcome
Returns the outcomes spread out in population as a list
"""
uniform_p = 1/len(Y)
print(uniform_p)
total_pops = []
for i in range(0,len(Y)):
each_o = [i]*(int(uniform_p*N))
total_pops += each_o
shuffle(total_pops)
return total_pops


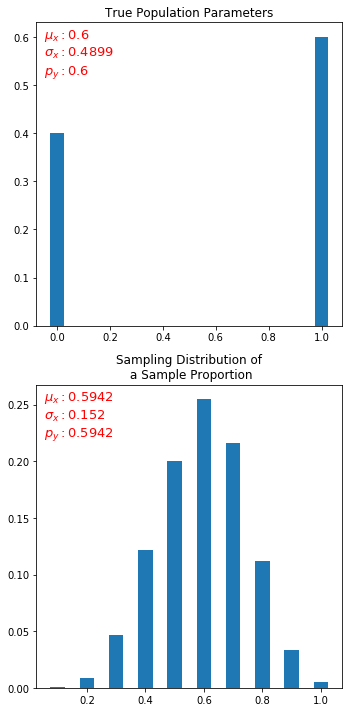
can you please share your matplotlib settings? I think you have the plot truncated, you are correct in that the sample distribution of the sample proportion on a bernoulli should be normally distributed around the population expected value ...
perhaps using something as:
to check if there are no graph issues